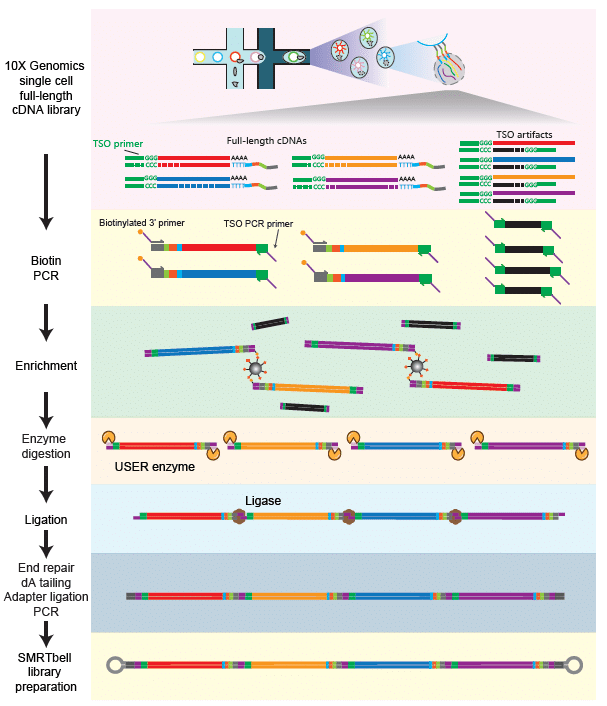
Long Read Single Cell Sequencing
Long Read Single Cell Sequencing - Web for ont sequencing, a r9.4.1 flow cell was used with a minion. Finally, on a perturbation experiment for 9 splicing factors, longcell identified regulatory targets that are validated by targeted sequencing. However, it is inadequate for comprehensive characterization of rna isoforms. Ont sequencing is one of the leading sequencing technologies in plant genomics , and was applied in this project to generate a complete plastome sequence based on long reads. Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations and gene expression profiles at the single cell level. In this review, we introduce. Scnanogps provides a robust computational tool to achieve this goal. Submersum sequencing, the flow cell. Web overall, our results demonstrate that single cell long read sequencing, in conjunction with short read sequencing, provides a robust method for comprehensively characterizing mrna sequences at the single cell level and identifying isoforms involved in the regulation of specific plasmodium stages. Web on matched single cell and visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, longcell found concordant signals between the two data modalities.
Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations and gene expression profiles at the single cell level. In this review, we introduce. This has several inherent advantages. Web on matched single cell and visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, longcell found concordant signals between the two data modalities. Web overall, our results demonstrate that single cell long read sequencing, in conjunction with short read sequencing, provides a robust method for comprehensively characterizing mrna sequences at the single cell level and identifying isoforms involved in the regulation of specific plasmodium stages. Scnanogps provides a robust computational tool to achieve this goal. Submersum sequencing, the flow cell. However, it is inadequate for comprehensive characterization of rna isoforms. Web for ont sequencing, a r9.4.1 flow cell was used with a minion. Finally, on a perturbation experiment for 9 splicing factors, longcell identified regulatory targets that are validated by targeted sequencing.
This has several inherent advantages. Submersum sequencing, the flow cell. Web for ont sequencing, a r9.4.1 flow cell was used with a minion. Ont sequencing is one of the leading sequencing technologies in plant genomics , and was applied in this project to generate a complete plastome sequence based on long reads. Web on matched single cell and visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, longcell found concordant signals between the two data modalities. Finally, on a perturbation experiment for 9 splicing factors, longcell identified regulatory targets that are validated by targeted sequencing. However, it is inadequate for comprehensive characterization of rna isoforms. Web overall, our results demonstrate that single cell long read sequencing, in conjunction with short read sequencing, provides a robust method for comprehensively characterizing mrna sequences at the single cell level and identifying isoforms involved in the regulation of specific plasmodium stages. (c) insertions and deletions from a single. Scnanogps provides a robust computational tool to achieve this goal.
SingleCell RNA Sequencing PacBio
Submersum sequencing, the flow cell. In this review, we introduce. Web on matched single cell and visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, longcell found concordant signals between the two data modalities. (c) insertions and deletions from a single. Web overall, our results demonstrate that single cell long read sequencing, in conjunction with.
Highthroughput targeted longread single cell sequencing reveals the
In this review, we introduce. Web for ont sequencing, a r9.4.1 flow cell was used with a minion. However, it is inadequate for comprehensive characterization of rna isoforms. Submersum sequencing, the flow cell. Web on matched single cell and visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, longcell found concordant signals between the two.
Shortread and targeted longread single cell sequencing of
Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations and gene expression profiles at the single cell level. Web for ont sequencing, a r9.4.1 flow cell was used with a minion. Scnanogps provides a robust computational tool to achieve this goal. This has several inherent advantages. Web on.
Major shortread and longread sequencing technologies. (A) Illumina
Ont sequencing is one of the leading sequencing technologies in plant genomics , and was applied in this project to generate a complete plastome sequence based on long reads. This has several inherent advantages. Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations and gene expression profiles at.
Highthroughput targeted longread single cell sequencing reveals the
Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations and gene expression profiles at the single cell level. (c) insertions and deletions from a single. Web for ont sequencing, a r9.4.1 flow cell was used with a minion. Ont sequencing is one of the leading sequencing technologies in.
Highthroughput targeted longread single cell sequencing reveals the
In this review, we introduce. Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations and gene expression profiles at the single cell level. Submersum sequencing, the flow cell. (c) insertions and deletions from a single. Web overall, our results demonstrate that single cell long read sequencing, in conjunction.
Nanopore LongRead RNAseq Reveals Widespread Transcriptional Variation
Web on matched single cell and visium long read sequencing for a tissue of colorectal cancer metastasis to the liver, longcell found concordant signals between the two data modalities. However, it is inadequate for comprehensive characterization of rna isoforms. Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia, the diversity of cell populations.
Singlecell RNA sequencing. (A) DropSEQ workflow, as described in
Scnanogps provides a robust computational tool to achieve this goal. (c) insertions and deletions from a single. Finally, on a perturbation experiment for 9 splicing factors, longcell identified regulatory targets that are validated by targeted sequencing. Ont sequencing is one of the leading sequencing technologies in plant genomics , and was applied in this project to generate a complete plastome.
Massively parallel longread sequencing of single cell RNA isoforms
Ont sequencing is one of the leading sequencing technologies in plant genomics , and was applied in this project to generate a complete plastome sequence based on long reads. However, it is inadequate for comprehensive characterization of rna isoforms. Submersum sequencing, the flow cell. Web although the lung is one of the most important organs mediating physiological adaptations to hypoxia,.
SingleCell Sequencing. SCS can be divided in three major steps (i
This has several inherent advantages. In this review, we introduce. Web overall, our results demonstrate that single cell long read sequencing, in conjunction with short read sequencing, provides a robust method for comprehensively characterizing mrna sequences at the single cell level and identifying isoforms involved in the regulation of specific plasmodium stages. Web although the lung is one of the.
Web On Matched Single Cell And Visium Long Read Sequencing For A Tissue Of Colorectal Cancer Metastasis To The Liver, Longcell Found Concordant Signals Between The Two Data Modalities.
In this review, we introduce. Scnanogps provides a robust computational tool to achieve this goal. Web overall, our results demonstrate that single cell long read sequencing, in conjunction with short read sequencing, provides a robust method for comprehensively characterizing mrna sequences at the single cell level and identifying isoforms involved in the regulation of specific plasmodium stages. Web for ont sequencing, a r9.4.1 flow cell was used with a minion.
Web Although The Lung Is One Of The Most Important Organs Mediating Physiological Adaptations To Hypoxia, The Diversity Of Cell Populations And Gene Expression Profiles At The Single Cell Level.
Submersum sequencing, the flow cell. However, it is inadequate for comprehensive characterization of rna isoforms. (c) insertions and deletions from a single. This has several inherent advantages.
Ont Sequencing Is One Of The Leading Sequencing Technologies In Plant Genomics , And Was Applied In This Project To Generate A Complete Plastome Sequence Based On Long Reads.
Finally, on a perturbation experiment for 9 splicing factors, longcell identified regulatory targets that are validated by targeted sequencing.